
Our genes provide the instructions to build our bodies, but errors or mutations in those instructions can have devastating effects. Pinpointing which of the millions of possible genetic mutations cause disease has remained one of the greatest challenges in human genetics. This knowledge is crucial to faster diagnosis and developing life-saving treatments.
Now, researchers at Google DeepMind have developed a groundbreaking AI system named AlphaMissense that provides a comprehensive catalog classifying the effects of nearly every possible human genetic mutation. As described in a paper published in Science, AlphaMissense achieved state-of-the-art performance at predicting mutation effects without being trained on human-curated variant databases.
This new tool has the potential to accelerate diagnostics, illuminate biological mechanisms underlying diseases, and pave the way toward new discoveries.
AlphaMissense focuses on a type of genetic mutation known as a missense variant, where a single "letter" change in the DNA sequence results in a different amino acid building block in the resulting protein. These subtle changes can severely disrupt a protein's structure and function, leading to genetic diseases like cystic fibrosis.
Out of over 71 million possible missense variants in the human genome, AlphaMissense was able to confidently classify 89% of them as either likely pathogenic or likely benign. This represents a major expansion in knowledge, considering only 0.1% of missense variants have been confirmed by human experts so far.
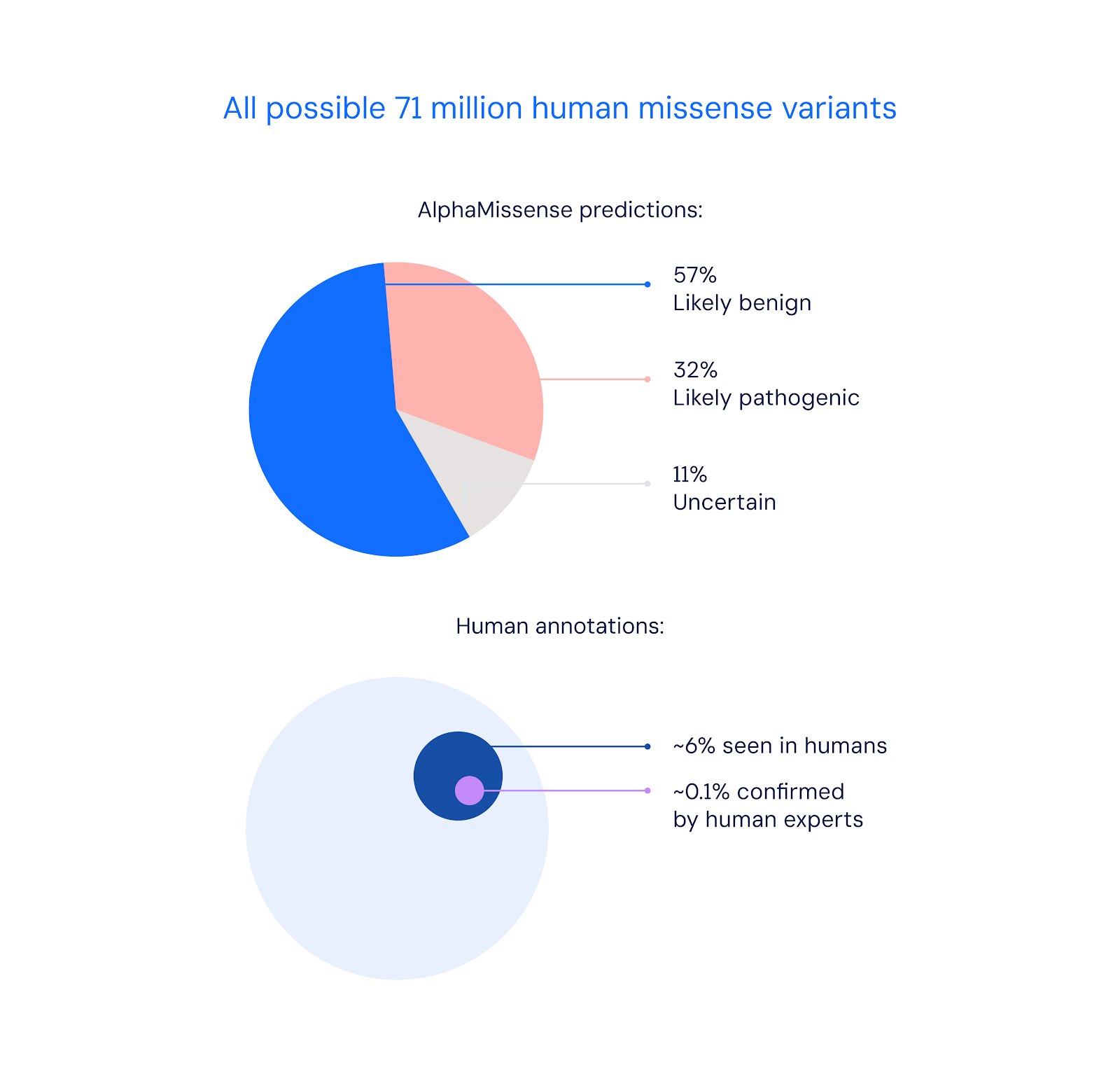
AlphaMissense achieved this through an AI architecture that combines evolutionary analysis of protein family sequences with the structural modeling capabilities of Google DeepMind's breakthrough protein folding model AlphaFold. The system was fine-tuned using weak labels derived from population frequency data, rather than direct human-curated clinical databases. This avoids inherent biases and allows more reliable evaluation across diverse datasets.
AlphaMissense also takes a unique approach in that it doesn't predict the mutation's effect on the protein structure or other effects on protein stability. Instead, it utilizes a vast array of related protein sequences and the structural context of variants, producing a score approximating the likelihood of a variant's pathogenic nature. This nuanced, continuous score allows researchers to set their desired accuracy parameters when categorizing variants.
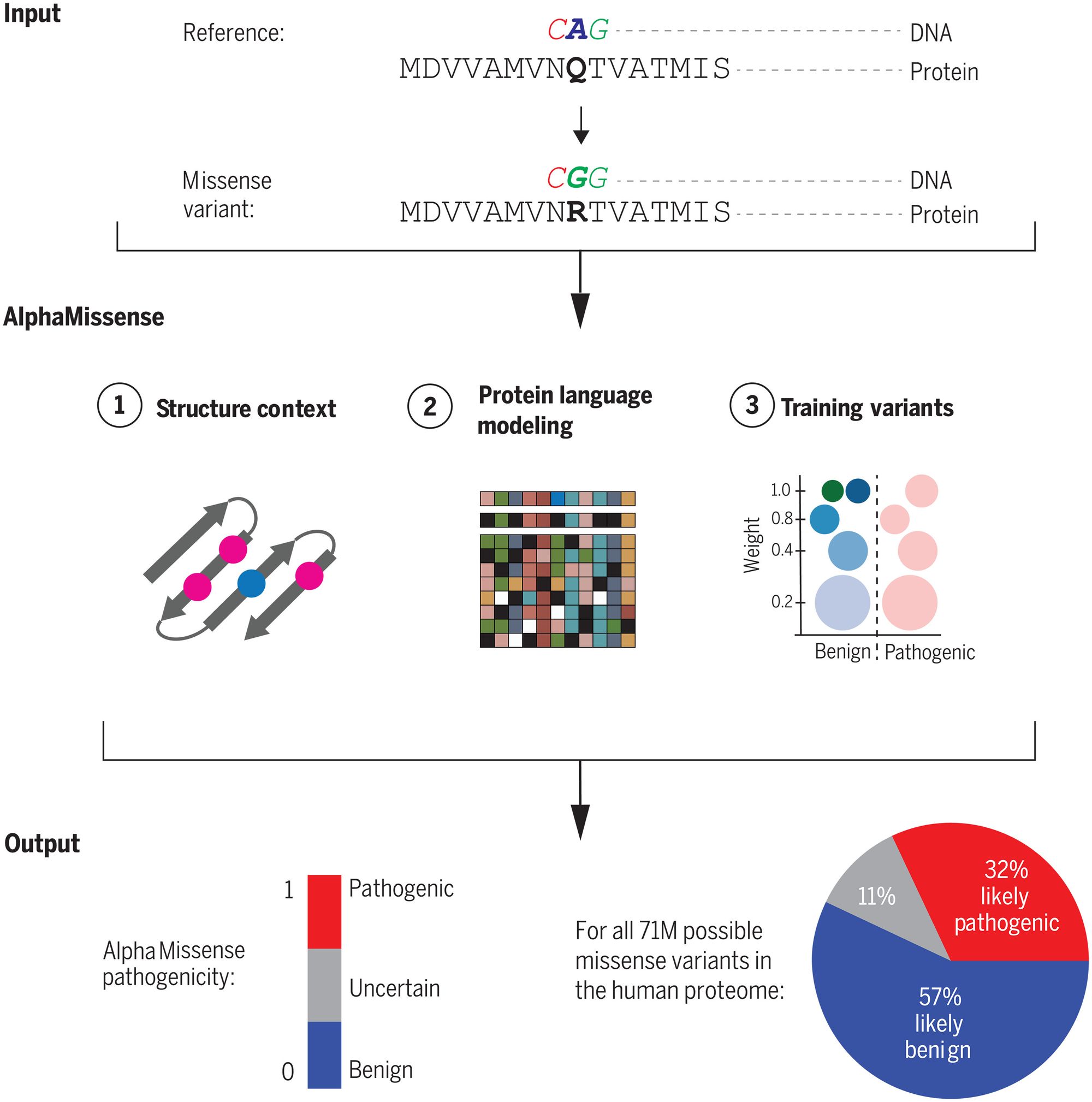
Remarkably, AlphaMissense reached state-of-the-art performance on clinically curated variant databases, experimental fitness assays, and assessments of de novo mutations underlying rare developmental disorders. Yet it achieved this accuracy without ever being directly trained on such gold-standard variant effect data. This demonstrates the model’s ability to capture intrinsic principles governing mutation effects on human health, enabling it to generalize beyond available training data.
Google must be applauded for open-sourcing the AlphaMissense model code and predictions dataset. A tool like this that can accurately classify the effects of mutations at scale has tremendous implications across biology and medicine. AlphaMissense provides a knowledge base that could significantly accelerate future discoveries.
For one, experimental biologists currently have to design laborious assays to test mutations in the lab. AlphaMissense’s predictions provide helpful insights that could prioritize the most informative mutations to study. Testing thousands of mutations across entire protein families in parallel could help reveal shared mechanisms underlying genetic diseases.
While these predictions aren't primed for direct clinical application and must be paired with other evidence sources, the possibilities they unlock are immense. From the rapid diagnosis of rare genetic disorders to identifying previously unknown disease-causing genes, the ramifications are profound. It also expands the scope for new gene discovery by enabling analysis of more genes in association studies of patients with undiagnosed conditions.
The scale of AlphaMissense enables entirely new investigations into the link between genotype and phenotype across the human proteome. Researchers can use these comprehensive predictions to uncover new disease-gene associations and better understand the biological impacts of mutations in their genetic studies.
In a world where genetic disorders remain enigmatic and elusive, tools like AlphaMissense herald a hopeful, informed future. Only time will tell the true magnitude of its impact, but for now, it stands as a beacon of promise in the vast, intricate world of human genetics and exemplifies the transformative potential of AI.

